Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
Carousel with three slides shown at a time. Use the Previous and Next buttons to navigate three slides at a time, or the slide dot buttons at the end to jump three slides at a time. CHLORIDE Cl

Ketan D. Patel, Philip B. d'Andrea, ... Andrew M. Gulick
Guiyang Yao, Simone Kosol, … Roderich D. Süssmuth
Jie-Ren Deng, Nathanael Chun-Him Lai, … Man-Kin Wong
Ahmad Altiti, Mingzhu He, … Yousef Al-Abed
James R. Marshall, Peiyuan Yao, … Nicholas J. Turner
Kohei Umedera, Atsushi Yoshimori, … Hiroyuki Nakamura
Jian Tang, Hongfei Chen, … Huan Wang
Zhuozhuo He, Lingzi Peng & Chang Guo
Shay Laps, Fatima Atamleh, … Ashraf Brik
Scientific Reports volume 6, Article number: 22856 (2016 ) Cite this article
Herein we present the synthesis of a novel type of peptidomimetics composed of repeating diaminopropionic acid residues modified with structurally diverse heterobifunctional polyethylene glycol chains (abbreviated as DAPEG). Based on the developed compounds, a library of fluorogenic substrates was synthesized. Further library deconvolution towards human neutrophil serine protease 4 (NSP4) yielded highly sensitive and selective internally quenched peptidomimetic substrates. In silico analysis of the obtained peptidomimetics revealed the presence of an interaction network with distant subsites located on the enzyme surface.
Several useful methods to generate structurally diverse substrates to study the specificity of proteolytic enzymes currently exist1. In general, these substrates are short peptides containing a reporter chromogenic, fluorogenic or luminogenic group2. Specific peptide sequences are usually based on compounds naturally interacting with proteases and are further refined using enzyme crystal structure analysis as well as in silico studies3. Alternatively, high throughput screening of libraries composed of peptides or peptidomimetics yields optimized substrate recognition sequences4. This method has been routinely applied for the determination of protease specificity and the development of peptide-based protease probes5. A major limitation of this approach is the overlapping specificities of different proteases6. Closely related proteases with similar substrate recognition patterns are able to cleave similar peptide sequences at equivalent rates. In order to enhance the selectivity of such peptides, their original sequences are usually modified by replacing some of the proteinogenic amino acid residues with structural synthetic analogs7. Recently, Kasperkiewicz et al.8 described the development of a substrate library composed of non-proteinogenic amino acid residues. This library was validated against human neutrophil elastase and yielded a very potent fluorogenic substrate which was further converted into a specific activity-based probe.
Herein, we propose a novel and universal method to design and synthesize a library of peptide-like compounds composed of a series of diaminopropionic acid (Dap) residues substituted by heterobifunctional polyethylene glycol moieties. This library contains a Dap residue substituted on the amino group by a set of polyethylene glycol moieties at positions P2–P4, while the P1 position is fixed with a particular amino acid residue (in this case of human neutrophil serine protease 4 the P1 position is occupied by an Arg residue).
Our goal was to cover a broad range of chemical interactions between the enzyme and its substrate molecule. In each position, the chemical groups differed by side chain character specificity including amino, guanidino, carboxy, methoxy, hydroxy and carbobenzyloxy groups (basic, acidic, neutral, hydrophilic, hydrophobic) which was provided via the variety of functionalized PEG derivatives. Simultaneously, the effect of side chain length was investigated since three possibilities of length for each side chain were available: first, simple amino acid chain or its stable protected form; second, side chain extended by single ethylene unit form of all components; third, replication of chemical entities with two polyethylene unit extension. Importantly, the introduction of PEG residues made the prepared libraries highly soluble in water.
This approach guaranteed the selection of the optimal residue at each position (Fig. 1). Diversifying the library by inserting 18 different compounds at every X2–X4 position led to the generation of almost 6000 compounds covering a broad range of chemical space. The library was synthesized using manual solid-phase synthesis applying Fmoc chemistry and was screened using an iterative approach in solution. The release of ANB-NH2 quencher resulted in an increase of free ABZ-peptide fluorescence that allowed us to determine the optimal residue at each examined position. Incorporation of polyethylene glycol residue to the protein or peptide (selective PEGylation) resulted in improved water solubility and higher proteolytic stability of modified molecules; while increasing the molecular weight prolonged the half-life of conjugates9,10. Selective PEGylation has been successfully used for cross-linking of molecules11, synthesis of peptide-like polymers modified with different chemical entities12, generation of self-assembling nanostructures13, cell delivery of molecules and many other applications (described in ref. 14).
Overview of library and its components.
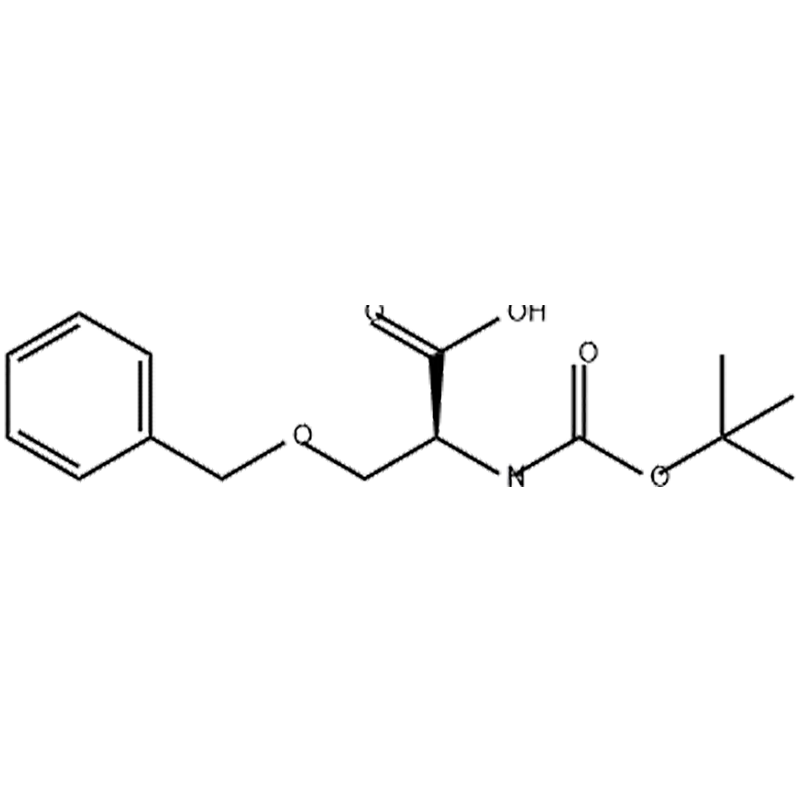
5-(tert-butyloxycarbonyl-amino)-3-oxapentanoic acid (further abbreviated as O1), 8-(tert-butyloxycarbonyl-amino)-3,6-dioxaoctanoic acid (O2), 5-[N-tert-butyloxycarbonyl-N′-(2,2,4,6,7-pentamethyldihydro benzofuran-5-sulfonyl)]amidino-3-oxapentanoic acid (GO1), 8-[N-tert-butyloxycarbonyl-N′-(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl)]amidino-3,6-dioxaoctanoic acid (GO2), 5-(benzyloxycarbonyl-amino)-3-oxa-pentanoic acid (CbzO1), 8-(benzyloxycarbonyl-amino)-3,6-dioxaoctanoic acid dicyclohexylamine (CbzO2), 2-(2-tert-butoxyethoxy) acetic acid (HO1), 2-(2-(2-tert-butoxyethoxy)ethoxy) acetic acid (HO2), 3,6-dioxaoctanedioic acid 1-tert-butyl ester (CO1), 3,6,9-trioxaundecandioic acid 1-tert-butyl ester (CO2), 5-methoxy-3-oxapentanoic acid (MO1), 8-methoxy-3,6-dioxaoctanoic acid (MO2) along with l-2,3-diaminopropionic acid Dap, Cbz-protected Dap, Ser, Ser methyl ether, l-2-amino-3-guanidino-propionic acid (Agp) and Asp.
Our target enzyme in this study was human neutrophil serine protease 4 (NSP4), recently discovered by Jenne et al.15,16. The amino acid sequence of NSP4 shares almost 39% identity with the sequences of human neutrophil elastase (HNE) and proteinase 3 (PR3). NSP4, together with HNE, cathepsin G (CatG) and PR3, is released into the pericellular environment as a result of neutrophil activation. Similar to other neutrophil serine proteases (NSPs), NSP4 is synthesized as an inactive precursor (preproNSP4) which, after the removal of signal peptide, is activated by cathepsin C. In contrast to other NSPs, NSP4 displays a strong preference towards an arginine residue at the P1 position of the substrate (according to Schechter-Berger nomenclature17), but its primary sequence forecasts a very different elastase-like active site with a preference for small aliphatic amino acids. This complication was recently unraveled by Lin et al. who solved the crystal structure of NSP4 complexed with d -Phe-l -Phe-l -Arg chloromethyl ketone18. Based on the provided data, the Arg residue is in a non-canonical “up” form stabilized by a network of hydrogen bonds involving the guanidinium group and Ser residue of the enzyme and hydrophobic shallow cleft that interacts with the aliphatic side chain of the Arg typical for elastase-like proteases. Due to this atypical substrate binding mode, NSP4 is able to recognize and process substrates containing arginine-like residues such as citrulline or methylated arginine.
In order to construct a novel type of internally quenched library, we used different monoprotected heterobifunctional PEG moieties to diversify a library based on a diaminopropionic acid scaffold (Fig. 1 presents the library formula and its chemical components). The library was synthesized using the mix and split method via Fmoc chemistry. The formation of Dap-PEG conjugates was performed on resin via a submonomeric method (one example of DAPEG-based molecule synthesis is presented in Fig. 2). Our goal was to investigate P2, P3 and P4 subsite preferences with a fixed P1 position occupied by an Arg residue. Each peptide in the library was flanked by a C-terminal amide of 5-amino-2-nitrobenozic acid (acceptor of fluorescence) and an N-terminal 2-amino benzoic acid (fluorescence donor) pair which allowed for simultaneous UV and fluorescence monitoring. Screening process was performed using an iterative mode of deconvolution in solution.
An example of a synthetic scheme for DAPEG-based peptidomimetic preparation.
The 18 sublibraries with fixed X4 positions were incubated with NSP4. The highest hydrolysis rate of the Arg-ANB-NH2 bond was observed for diaminopropionic acid modified by carbobenzyloxy-protected amino polyethylene glycols Dap(O2(Cbz)) and Dap(O1(Cbz) (Fig. 3A) followed by the remaining amino acid residues - Dap and the Cbz-protected analog (Dap(Cbz)) and methyl ether of Ser. A relatively high rate of proteolysis was also observed of methyl ether derived PEG chains with supremacy of the longer Dap(MO2) chain - Insignificant output was observed for acidic residues despite side chain length. Considering the above findings, in next our step deconvolution of Dap(O2(Cbz)) was included yielding the ABZ-Dap(O2(Cbz))-X3-X2-Arg-ANB-NH2 library.
The deconvolution of the library (at average concentration 3.4 × 10−5 M) with general formula ABZ-X4-X3-X2-Arg-ANB-NH2 against NSP4 (1.46 × 10−8 M).
Next, 18 sublibraries with randomized X3 position were analyzed. In assayed position, the most intense proteolysis was observed for compounds with short side chain amino acid residues - Dap(Cbz), Dap and Agp (Fig. 3B). The highest rate of hydrolysis was observed for the residue with aromatic ring present on its side chain - Dap(Cbz). Other residues were poorly processed by NSP4 with 3–4 times lower hydrolysis rate. Importantly, the proteolytic efficacy of the best sublibrary was three times greater than the most potent residue found for the X4 position. Following analysis, Dap(Cbz) was included at position X3 resulting in an 18-member library with a general formula of ABZ-Dap(O2(Cbz))-Dap(Cbz)-X2-Arg-ANB-NH2.
Incubation of each of 18 peptides modified at position X2 with NSP4 indicated that Dap analogs substituted by PEG chains displayed the highest fluorescent intensity (Fig. 3C). Within this set, the PEG chain with guanidinium group was processed at the highest rate. Low proteolysis was observed for the set of short-chained amino acid - Dap, Dap(Cbz), Agp or Asp with the only exception being Ser and its methyl ether. The sequence ABZ-Dap(O2(Cbz))-Dap(Cbz)-Dap(GO1)-Arg-ANB-NH2 (further abbreviated as substrate 1) was selected as most susceptible for NSP4-mediated proteolysis.
A mass spectrometry quality check was performed at all stages of library deconvolution and all library components were identified within the HR MS spectra (Fig. S1). Similar analyses were performed for randomly selected sublibraries for each assayed position (X4, X3 and X2) identifying all signals corresponding to [MW+H]+ or its sodium/potassium adducts (Figs S9, S10 and S11).
The obtained substrate 1 was characterized in order to verify its chemical composition and sequence. The HPLC and MS analysis of substrate 1 indicated a purity >98% and a molecular weight equal to 1271.34. Formation of a well-defined y-ion series was observed following MS/MS fragmentation (see Figs S2 and S3) and these, together with the ions originating from Cbz loss/cleavage fully validated the structure of substrate 1. The structure of substrate 1 was further validated using the 2D 1HNMR method (details are provided in Supplementary Material, Figs S4, S5 and S6).
Incubation of substrate 1 (tR = 24.96, MW 1271.34) with hNSP4 (Fig. 4) resulted in cleavage of the peptide bond located between Arg and ANB-NH2. The fluorescent fragment, ABZ-Dap(O2(Cbz))-Dap(Cbz)-Dap(GO1)-Arg-OH (tR = 23.46, MW 1108.21), lacking its C-terminal quencher group, was no further degraded up to 48 h under the conditions of our assay (Fig. 4).
HPLC analysis of substrate 1 (1.97 μM) incubated with hNSP4 (14 nM) at different time points: 0, 15 min, 24 h and 48 h.
The obtained substrate 1 displayed high affinity (expressed as KM reaching 7.6 × 10−6 M) and moderate kcat value of 1 s−1 and its specificity parameter value reached 1.3 × 105 M−1 × s−1 (Fig. S7).
The incubation of substrate 1 with neutrophil serine proteinases (CatG, HNE, or PR3) at a concentration of 10−8 M produced no visible proteolysis (see Fig. 5A). Analogous experiment with set of proteases related to blood cells, including cathepsins and kallikreins, revealed that the developed substrate is susceptible to NSP4-mediated proteolysis and to a much lesser extend to degradation by KLK14. No other proteolytic enzyme at a concentration of 10−8 M was able to cleave any of the peptide bonds of probe 1 at a noticeable level (Fig. 5B).
Incubation of substrate 1 (1.97 μM) with (A) neutrophil serine proteases (cathepsin G, elastase and proteinase 3) at a concentration of 8.7 nM; (B) set of blood-associated proteases involving cathepsins and kallikreins.
The incubation of substrate 1 with decreasing amounts of hNSP4 yielded a visible fluorescence (signal-to-noise ratio of 3:1) at 10−11 M of the studied enzyme (Fig. S8).
A molecular docking approach was employed to investigate the substrate 1-NSP4 binding mode. The most probable ligand-protein complex is presented in Fig. 6. The ANB residue is located near His41 and Arg62 and is able to create two types of interactions: π-π between the ANB and His41 aromatic rings and two hydrogen bonds between the amine group of ANB and the carbonyl oxygen of His41 and between nitro group of ANB and Arg62. The arginine residue in the X1 position is located in the Ser214 binding pocket and is stabilized by two hydrogen bonds with Ser216 and the backbone of Gly217. Additionally, it can form a cation – π interaction with the Phe190 residue. According to experimental data, the preferred residue in the X2 position is Dap(GO1). Our model shows that this residue is located in the cavity normally occupied by the substrate backbone. The guanidinium group of Dap(GO1) is located in the nest created by the side chains of His99, Trp172, His175 and Phe215 (S4 pocket), thus creating strong cation – π interactions. Additionally, we have found a hydrogen bond with the backbone of Trp172. The peptide bond between arginine (X1) and ANB-NH2 is located in the proximity of the Ser195 found within the catalytic triad in an orientation which allows for a nucleophilic attack at the carbonyl carbon. The most preferred residue in the X3 position is Dap(Cbz). It is located near the His57 residue of the catalytic triad and in this case we found backbone-backbone interactions. Additionally, we have found a cation – π interaction with Arg62. The most preferred residue in the X4 position is Dap(O2(Cbz)). The chain Dap(O2(Cbz)) is located in the vicinity of His61, however the phenyl ring of the carbobenzyloxy moiety is located in the nest created by Phe59, Leu88 and Thr90. We did not find any significant interactions of the ABZ residue with NSP4.
Proposed putative model of substrate 1 binding to the active site of NSP4 (based on 4Q7Z.pdb).
The enzyme binding-site is presented as surface with the residues composing the binding pocket and catalytic triad (Ser195 – orange, His57 – blue, Asp102 - red) represented as sticks.
Proteases belong to one of the largest group of enzymes and have a broad range of activity and overlapping cleavage sequences/specificities. Chasing a single proteolytic enzyme in complex biological systems is a demanding challenge. In this work, the synthetic method used for the development of the library represents novel atypical peptidomimetics based on a submonomeric approach. The presence of a fluorogenic ABZ (2-amino benzoic acid) moiety and ANB-NH2 (amide of 5-amino-2-nitrobenzoic acid) fluorescence quencher pair facilitates in-solution fluorescence screening. The introduction of novel pseudo amino acid side chains such as those functionalized by different chemical entities of polyethylene glycol moieties covers a broad range of interactions between the enzyme studied and the selected substrate. The sequence revealed upon deconvolution of such DAPEG (Dap modified by PEG moiety) library supplement by typical amino acid residues is a novel approach, lacking any similarity to any work reported recently by our group. It has been constructed with two DAPEG units with different functional group on the side chains. Such diversified functional groups together with their glycol side chains offer multiple contacts between the enzyme and the selected substrate. As indicated, in position X4 a long polyethylene glycol side chain terminated by an aromatic group is preferred over other chemical entities at this position. In general, this result is in agreement with data provided by Perera et al.15 since NSP4 prefers an aromatic amino acid in this position. However, the side chain length is an important factor that makes a significant difference between Phe/Tyr and Dap(O2(Cbz)). In the light of data summarized in Table 1 that provides published up to date NSP4 substrate sequences, position P4 is dominated by hydrophobic amino acid residues (hCha, Phe).
Positions X3 and X2 are also occupied by atypical residues; however, the short length of Dap(Cbz) in the X3 position could be considered an aromatic amino acid imitation since Dap(GO1) in X2 structurally resembles homologous elongated form of Arg. As presented in Table 1, combinations of amino acid residues with hydrophobic and basic side chains (Ile/Arg, Phe(4-guanidine)/Oic, Lys/Pro) are preferred in the P2/P3 positions. A similar pattern is observed in our newly developed substrate where Dap(Cbz) in position P3 is followed by pegylated analog of Arg (Dap(GO1) in position P2.
The kinetic parameters, especially kcat/KM ratio (over 1.31 × 105 M−1 × s−1) of the developed compound are superior over other NSP4 substrates reported thus far (summarized in Table 2) and display specificity 10-fold greater than a substrate described by Lin et al.18 and 4-times higher than a substrate selected by Kasperkiewicz et al.19
The presence of factionalized glycol side chains in the Dap scaffold of these peptidomimetics offers high selectivity over other proteolytic enzymes. This was demonstrated using a panel of over 20 neutrophil-related proteinases which included NSPs and a family of human kallikreins recently found to be expressed in neutrophils (Fig. 5)20. The kinetic parameters of substrate 1 and together with its low (subnanomolar) detection limit make this peptidomimetic an excellent tool to study NSP4 activity in biological material. Until now, there is a limited set of data regarding the NSP4 specificity. According to Perera et al., the non-primed segment (spanning residues P4 - P1) of the peptide chain interacting with NSP4 should be composed as follows: Gly-Ile-Pro-Arg. The substrate sequence obtained in this study did not display any obvious similarity, except for the Arg residue in the P1 position, which ensures the proper substrate recognition by the enzyme. The exceptional proteolytic stability of the cleaved N-terminal fragment of the peptidomimetic chain is worth highlighting, as this clearly indicates that the only peptide bond susceptible to NSP4-mediated hydrolysis is located between the Arg and amide of ANB. In the molecular docking model, the binding of the P1 arginine side chain is stabilized by the interactions of the guanidinium group with residues building the external surface of S1 pocket and the NH2 of ANB group forming a hydrogen bond with His41.
The low solubility of small peptidomimetics or modified peptides often hampers their use in well-designed experiments. This is not the case for this class of substituted Dap derivatives as exemplified by our compound 1 which displays excellent solubility in water.
The unnatural character and considerable size of the side chains of the presented NSP4 substrate (compound 1) together with its significant differences from previously reported NSP4 substrate specificity in the P4-P2 pockets (Gly-Ile-Pro) provoked us to investigate alternative modes of binding. According to Perera et al.16 arginine and serine residues located at the C-terminal part of the ligand display the highest activity toward NSP4. This result is consisted with the proposed model of binding. The arginine residue in the P1 position is long enough to create a hydrogen bond with Ser216 and the backbone of Gly217. A small residue such as ANB is able to create more hydrogen bonds (with His41 and Arg62) in comparison to serine in discussed position (P1′). Our results indicate that the most preferred residue in the P2 position is Dap(GO1) while Perera et al.16 suggest arginine residue in this position. With the proposed mode of binding Dap(GO1) is located in the cavity normally occupied by the substrate backbone. This cavity is highly hydrophobic, created by the following residues: His99, Trp172, His175 and Phe215 (S4 pocket). None of the investigated residues fits better to the described cavity than Dap(GO1). Dap(MO1) and Dap(MO2) can create a set of hydrophobic interactions, however those residues are not able to create any hydrogen bonds. Dap(O1), Dap(O2), Dap(HO1) and Dap(HO1) are able to create hydrogen bonds, however, due to their high conformational flexibility, they are not able to maintain stable and strong interactions. The side chain of the residue in the P2 position cannot be too long, since we did not observe significant activity with Dap(GO2). According to the proposed model of binding, the most preferred residue in the P3 position should be Dap(GO1) and Dap(GO2) since both residues are able to form a salt bridge with Arg62. However, our experimental results showed the opposite effect indicating that Dap(Cbz) was the most preferred residue at the P3 position. The residue at the P3 position is located adjacent to the catalytic triad (especially H57). It might be suspected that the role of the P3 residue is also to protect the catalytic triad from external influence. The most interesting result was obtained in regards to the P4 position which seems to be occupied by an aromatic residue. Moreover, the chain length seems to be important in the activation process: the short chain (Dap(Cbz)) would interact with His61 and/or Pro96 while its elongation would weaken this interaction as a slight decrease in the NSP4 activity with Dap(O1(Cbz)) was observed. When the side chain was long enough to reach the protein surface we observed a major increase in NSP4 activity. Hence, the proposed binding place of the P4 residue is located beyond the ligand binding site; thus position P4 could be considered as a target for the design of specific and highly selective ligands for the NSP4.
To our best knowledge, the presented approach is unique and could effectively be applied for the development of novel selective synthetic substrates of any proteolytic enzyme. Our preliminary data indicate that following this approach novel substrates with superior kinetic parameters could be obtained for other proteolytic enzymes with canonical substrate binding pockets such as trypsin or matriptase (unpublished data). Iterative screening of DAPEG librarys could be applied to any proteolytic enzyme with known primary specificity. Fixing the P1 residue assures primary specificity while introducing variability of chemical entities that differ by length, charge and hydrophobicity at other positions to yield the optimal substrate sequence.
The method proposed here enriches the panel of previously known methods used to investigate secondary binding sites (exosites) on the surface of proteases. So far, only the exosite cellular libraries of peptide substrates (eCLiPS) approach proposed by the Daugherty group deals with protease exosite identification21. In this work the cellular libraries of peptide substrates (CLiPS) method was modified to support the screening process of peptides that enhance the proteolysis rate of a model peptide substrate. Both the peptide substrate and a peptide library of exosite ligands were expressed on cell surface of E. coli. The eCLiPS approach characterizes the functions of protease exosites and recognizes peptide sequences that are crucial for boosting protease specificity for natural protein substrates. This is not the issue in the method developed by us since it instead yields the secondary specificity of a protease forming a contact with distant subsites.
This novel class of peptidomimetic offers practically unlimited possibilities for generation of new peptide-like molecules with diverse application not only limited to protease research. An enormous number of protecting groups that could be attached to functionalized polyethylene glycol moieties in combination with different glycol length creates an excellent chemical repertoire for the development of new kinds of molecules with diverse activities.
The synthesis of the ANB-based library or peptides was initiated by the deprotection of the amino groups of the resin with 20% piperidine in DMF and the coupling of 5-amino-2-nitrobenzoic acid using a mixture of N,N,N′,N′-tetramethyl-O-(benzotriazol-1-yl)uronium tetrafluoroborate (TBTU)/4-dimethylaminopyridine (DMAP). The resin was washed twice in N methylmorpholine. Next, 2 equiv. of ANB was dissolved in DMF and 2 equiv. of TBTU was added, followed by 1 equiv. of DMAP. The obtained solution was added to the resin and after 30 s, 4 equiv. of N,N-diisopropylethylamine (DIPEA) was added. The whole mixture was stirred for 3 h. The solution was filtered off and the resin was washed with DMF. The procedure was repeated three times. Next, the first amino acid residue was coupled with a method described in ref. 22. A ninefold excess was applied to the active resin sites as follows: the amino acid was dissolved in pyridine (10 ml pyridine to 1 g peptidyl-resin). The whole solution was mixed until the temperature of −15 °C was reached and then 9 equiv. of POCl3 was added. The mixture was successively stirred for 20 min at −15 °C, 20 min at room temperature and 6 h in an oil bath. After deprotection with 20% piperidine in DMF, the peptide chain was elongated as follows: resin was divided into 18 equal parts and in six positions, the amino acid derivatives such as Fmoc-Asp(tBu), Fmoc-Ser(OtBu), Fmoc-Ser(OMe), Fmoc-Dap(Cbz), Fmoc-Agp(Boc)2, Fmoc-Dap(Boc) were introduced—where Dap is (S) 2,3-diaminopropionic acid and Agp ((S)2-amino-3-guanidino-propionic acid)—using a standard Fmoc synthesis protocol. To each of remaining 12 portions, Fmoc-Dap(Mtt) was introduced and 4-methyltrityl (Mtt) was removed using procedure described in ref. 23 (2% TFA in DCM with addition of 1% triisopropylsilane). Such mixture was added to every 12 portions and stirred for 15 min. Completeness of Mtt removal was verified by adding pure TFA and yellow color development was monitored to indicate the presence of free Mtt groups. The whole procedure was repeated until no absorbency increase monitored at 410 nm was recorded. Next, DIPEA/DMF solution was added to each system for 15 min, resulting in 12 portions of Fmoc-Dap-Arg-ANB-polymer. To each of the above resin aliquots, the monoprotected heterobifunctional polyethylene glycol moieties (PEG) were coupled using equimolar amounts of PEG/DIPCI/HOBt in DMF/NMP (1:1, v/v) solution. The following protected PEG derivatives were used: 5-(t-butyloxycarbonyl-amino)-3-oxapentanoic acid (further abbreviated as O1), 8-(t-butyloxycarbonyl-amino)-3, 6-dioxaoctanoic acid (O2), 5-[N-t-butyloxycarbonyl-N′-(2,2,4,6,7-pentamethyldihydro benzofuran-5-sulfonyl)]amidino-3-oxapentanoic acid (GO1), 8-[N-t-butyloxycarbonyl-N′-(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl)]amidino-3,6-dioxaoctanoic acid (GO2), 5-(benzyloxycarbonyl-amino)-3-oxa-pentanoic acid (O1(Cbz)), 8-(benzyloxycarbonyl-amino)-3,6-dioxaoctanoic acid dicyclohexylamine (O2(Cbz)), 2-(2-tert-butoxyethoxy) acetic acid (HO1), 2-(2-(2-tert-butoxyethoxy)ethoxy) acetic acid (HO2), 3,6-dioxaoctanedioic acid 1-tert-butyl ester (C01), 3,6,9-Trioxaundecandioic acid 1-tert-butyl ester (CO2), 5-methoxy-3-oxapentanoic acid (MO1), 8-methoxy-3,6-dioxaoctanoic acid (MO2). Completeness of the coupling was monitored using a Kaiser test. Lack of free amino groups allowed us to move to the next step of library synthesis in which all resin portions were mixed and again divided into 18 parts. The whole procedure was repeated twice. Finally, the tert-butyloxycarbonyl derivative of 2-amino benzoic acid (Boc-ABZ-OH) was attached to the N-terminal amino group using the standard coupling method described above.
After completing the synthesis, the sublibraries or individual peptidomimetics were cleaved from the resin, using a TFA/phenol/triisopropylsilane/H2O mixture (88:5:2:5, v/v)24. The purity of the synthesized compounds and the correctness of the synthesis were confirmed using an RP-HPLC ChromNAV (Jasco, Japan) equipped with a Kromasil 100 C8 column (Knauer, Germany) equipped with a UV-Vis detector and a fluorescence detector. A linear gradient from 10% to 90% B within 45 min was applied (A: 0.1% TFA; B: 80% acetonitrile in A). The peptides were monitored at 226 nm. The molecular weights of the synthesized peptides and peptide libraries were confirmed by analysis of the mass spectra which were recorded on a Biflex III MALDI TOF mass spectrometer (Bruker Daltonics, Germany) using α-cyano-4-hydroxycinnamic acid as a matrix.
All NMR spectra were recorded in H2O/D2O (9:1 v:v) at 298 K using the Bruker AVANCE III 700 MHz (Bruker Daltonics, Bremen, Germany). The concentration of the sample was 2.12 × 10−4 M (0.7 ml). For the studied compound, the following spectra were recorded: 1D proton spectrum and 2D TOCSY and ROESY at mixing times of 80 ms and 200 ms as well as 1H-13C HSQC.
The peptide libraries were synthesized using a portioning-mixing method. Initially, 17.7 g of the solid support (TentaGel S RAM) was used for the first library, i.e. ABZ-X4-X3-X2-X1-ANB-NH2. A twofold molar excess of amino acid or appropriate monoprotected PEG derivative was used for the coupling. The other synthetic methods used were as described above.
For the enzymatic studies, we used human NSP4 expressed in a Pichia pastoris expression system as described in ref. 15. The NSP4 concentration used in the deconvolution of the library was 1.46 × 10−9 M. The bovine β-trypsin (Sigma Chem. Co., USA) concentration was determined by spectrophotometric titration with 4-nitrophenyl-4′-guanidinobenzoate (NPGB) at an enzyme concentration oscillating around 10−6 M. Standardized trypsin solution was used to titrate BPTI (Sigma Chem. Co., USA) which in turn served to determine the solution concentrations of the produced NSP4. The substrate ABZ-Met-Phe-Pro-Arg-ANB-NH2 recently developed in our lab was used. The deconvolution of peptide libraries was carried out by the iterative solution method25. The lyophilizates obtained for each sublibrary were dissolved in dimethyl sulfoxide (DMSO) to a final concentration of 5 mg/ml and then diluted tenfold in an assay buffer. The fluorescence tests were performed using a FluoroStar OMEGA fluorescent microplate reader (BMG, Germany). The excitation and emission wavelengths were 320 nm and 410 nm for the ABZ/ANB. Enzymatic hydrolysis of the peptide was performed in 20 mM Tris-Cl buffer supplemented by 150 mM NaCl (pH 7.4) at 37 °C and continued over 30 min.
All other enzymes (purchased from RD Systems, USA) were tested in concentration 2 × 10−9 M in 20 mM Tris-Cl buffer supplemented by 150 mM NaCl (pH 7.4) at 37 °C for 30 min. Same conditions as above were applied to the assay.
Assay conditions for the determination of the Michaelis constants (KM) and catalytic constants (kcat) were as noted above. The specificity constants (kcat/KM) were calculated from kcat and KM values. Measurements were performed with an enzyme concentration of 1.46 × 10−9 M. Three to five measurements were carried out for each compound (systematic error expressed as a standard deviation never exceeded 20%). The calculated initial hydrolysis rates were used as a measure of the substrate activity of the investigated peptides. All details of kinetic studies and the method of calculating kinetic parameters have been described elsewhere26.
For each sample of diluted substrate (concentrations 1.97 × 10−6 M), the appropriate amount of enzyme (1.46 × 10−8 M) was added and the solution was incubated for 5, 30 and 60 min. The progress of the proteolytic reaction was monitored by RP-HPLC. The individual peaks resulting from the hydrolysis reaction were collected and identified using the mass spectra which were recorded on a Biflex III MALDI TOF mass spectrometer (Bruker Daltonics, Germany) using α-cyano-4-hydroxycinnamic acid as a matrix.
The in silico study was performed using AutoDock Vina 1.1.2 molecular docking software27. For the optimization of compound 1 model geometry the MM2 force field was employed. The simulation of protein-substrate interaction was based on the crystal structure of NSP4 with inhibitor (Phe-Phe-Arg-chloromethylketone; 4Q7Z.pdb)18. Initially the inhibitor and water coordinates were removed from the protein structure and AutoDockTools 1.5.6 (Scripps Research Institute, USA) was employed to prepare the receptor and ligand models for docking studies. The molecular docking of flexible ligand to the rigid protein active center defined by the 50 × 50 × 20 Å grid box with a center at −14.5, −5.0, −1.7 was performed with the default settings.
How to cite this article: Wysocka, M. et al. PEGylated substrates of NSP4 protease: A tool to study protease specificity. Sci. Rep. 6, 22856; doi: 10.1038/srep22856 (2016).
Hu, H. Y. et al. FRET-based and other fluorescent proteinase probes. Biotechnol J. 9, 266–281 (2014).
Article CAS PubMed Google Scholar
Wysocka, M. & Lesner, A. Future of protease activity assays. Curr Pharm Des. 19, 1062–1067 (2013).
Article CAS PubMed Google Scholar
Korkmaz, B. et al. Design and use of highly specific substrates of neutrophil elastase and proteinase 3. Am J Respir Cell Mol Biol. 30, 801–807 (2004).
Article CAS PubMed Google Scholar
Schneider, E. L. & Craik, C. S. Positional scanning synthetic combinatorial libraries for substrate profiling. Methods Mol Biol. 539, 59–78 (2009).
Article CAS PubMed PubMed Central Google Scholar
Wysocka, M. et al. Substrate specificity and inhibitory study of human airway trypsin-like protease. Bioorg Med Chem. 18, 5504–5509 (2010).
Article CAS PubMed Google Scholar
Ziebuhr, J. et al. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J Virol. 81, 3922–3932 (2007).
Article CAS PubMed PubMed Central Google Scholar
Wysocka, M. et al. New chromogenic substrates of human neutrophil cathepsin G containing non-natural aromatic amino acid residues in position P(1) selected by combinatorial chemistry methods. Mol Divers. 11, 93–99 (2007).
Article CAS PubMed Google Scholar
Kasperkiewicz, P. et al. Design of ultrasensitive probes for human neutrophil elastase through hybrid combinatorial substrate library profiling. Proc Natl Acad Sci USA 111, 2518–2523 (2014).
Article CAS ADS PubMed PubMed Central Google Scholar
Ginn, C., Khalili, H., Lever, R. & Brocchini, S. PEGylation and its impact on the design of new protein-based medicines. Future Med Chem. 6, 1829–1846 (2014).
Article CAS PubMed Google Scholar
Veronese, F. M. & Mero, A. The impact of PEGylation on biological therapies. BioDrugs. 22, 315–329 (2008).
Article CAS PubMed Google Scholar
Lesner, A. et al. Fluorescent analogs of trypsin inhibitor SFTI-1 isolated from sunflower seeds-synthesis and applications. Biopolymers. 102, 124–135 (2014).
Article CAS PubMed Google Scholar
Gokhale, S., Xu, Y. & Joy, A. A library of multifunctional polyesters with “peptide-like” pendant functional groups. Biomacromolecules. 14, 2489–2493 (2013).
Article CAS PubMed Google Scholar
Sadatmousavi, P., Mamo, T. & Chen, P. Diethylene glycol functionalized self-assembling peptide nanofibers and their hydrophobic drug delivery potential. Acta Biomater. 8, 3241–3250 (2012).
Article CAS PubMed Google Scholar
Veronese, F. Peptide and protein PEGylation: a review of problems and solutions. Biomaterials 22, 405–417 (2001).
Article CAS PubMed Google Scholar
Perera, N. C. et al. NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proc Natl Acad Sci USA 109, 6229–6234 (2012).
Article CAS ADS PubMed PubMed Central Google Scholar
Perera, N. C. et al. NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity. J Immunol. 191, 2700–2707 (2013).
Article CAS PubMed Google Scholar
Schechter, I. & Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 (1967).
Article CAS PubMed Google Scholar
Lin, S. J., Dong, K. C., Eigenbrot, C., van Lookeren Campagne, M. & Kirchhofer, D. Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease. Structure. 22, 1333–1340 (2014).
Article CAS PubMed Google Scholar
Kasperkiewicz, P. et al. Design of a Selective Substrate and Activity Based Probe for Human Neutrophil Serine Protease 4. PLoS One. 10, e0132818 (2015).
Article PubMed PubMed Central Google Scholar
Jabaiah, A. M., Getz, J. A., Witkowski, W. A., Hardy, J. A. & Daugherty, P. S. Identification of protease exosite-interacting peptides that enhance substrate cleavage kinetics. Biol Chem. 393, 162–167 (2012).
Lizama, A. J. et al. Expression and bioregulation of the kallikrein-related peptidases family in the human neutrophil. Innate Immun. (2015) Jan 6. In press.
Hojo, K. et al.Chem.Pharm.Bull.(Tokyo) 48, 1740–1744 (2000).
Bourel , L. , Carion , O. , Gras-Masse , H. & Melnyk , O. The deprotection of Lys(Mtt) revisited.J Pept Sci.6, 264–270 (2000).
Article CAS PubMed Google Scholar
Sole, B. & Barany, G. Optimization of Solid-Phase Synthesis of. [Ala8]-dynorphin A. J. Org. Chem. 57, 5399–5403 (1992).
Houghten, R. A. et al. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature 354, 84–86 (1991).
Article CAS ADS PubMed Google Scholar
Wysocka, M. et al. Three wavelength substrate system of neutrophil serine proteinases. Anal. Chem. 84, 7241–7248 (2012).
Article CAS PubMed Google Scholar
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 31, 455–461 (2010).
CAS PubMed PubMed Central Google Scholar
This work was supported by National Science Center of Poland grant number UMO-2014/15/B/ST5/05311, UMO-2013/08/W/NZ1/00696 and Polish Ministry of Science and Higher Education Iuventus PLUS under grant IP2012 0596 72 (MW). The authors would like to thank Dr Keri Csencsits-Smith (University of Texas at Houston) for critical reading of the manuscript.
Faculty of Chemistry, University of Gdansk, Gdansk, Poland
Magdalena Wysocka, Natalia Gruba, Artur Giełdoń, Krzysztof Brzozowski, Krzysztof Rolka & Adam Lesner
Faculty of Chemistry, Wroclaw Technical University, Wroclaw, Poland
Renata Grzywa & Marcin Sieńczyk
Comprehensive Pneumology Center, Institute of Lung Biology and Disease, Helmholtz Zentrum München, Munich, Germany
Faculty of Chemistry, University of Wroclaw, Wroclaw, Poland
Remigiusz Bąchor & Zbigniew Szewczuk
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
M.W. and A.L. planned and designed the experiments, M.W., N.G., R.B., Z.S. and K.B. performed experiments, M.W. prepared Figures 1–5, R.G. prepared Figure 6, R.G., A.G., M.S. performed molecular dynamic study, R.B. and Z.S. performed mass spectrometry study, K.B. performed NMR study, A.L. and M.W. wrote the main manuscript, M.W. and A.L. prepared supporting information file. M.W., M.S., A.G., E.G., K.R., D.J. and A.L. edit and review the manuscript.
The authors declare no competing financial interests.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Wysocka, M., Gruba, N., Grzywa, R. et al. PEGylated substrates of NSP4 protease: A tool to study protease specificity. Sci Rep 6, 22856 (2016). https://doi.org/10.1038/srep22856
DOI: https://doi.org/10.1038/srep22856
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Scientific Reports (Sci Rep) ISSN 2045-2322 (online)
Schiff Base Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.